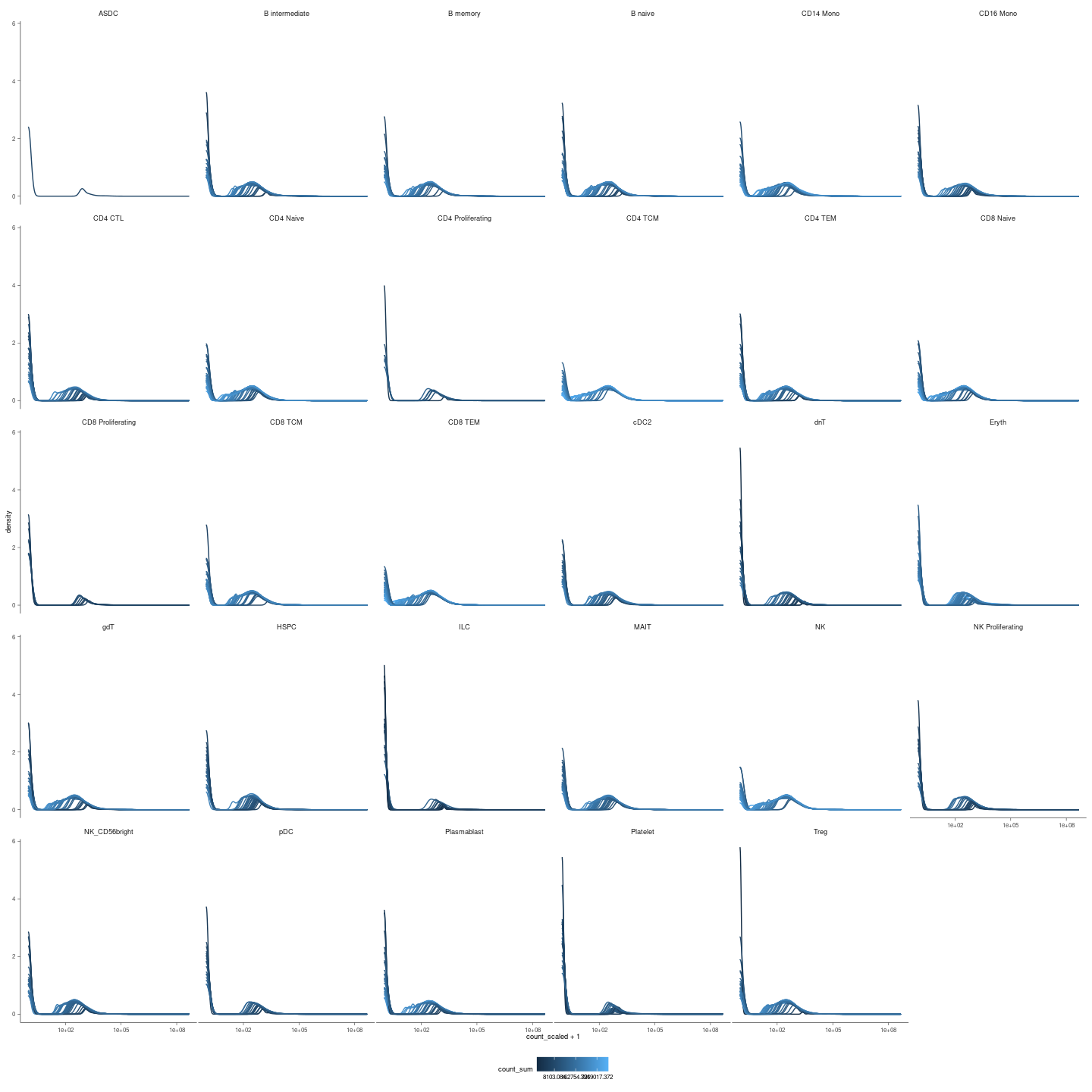
plots_density = readRDS(here("data/3_prime_batch_1/fast_pipeline_results/differential_transcript_abundance/plot_densities.rds"))
Density plots to confirm that scaling for seqwuencing depth worked
plots_density
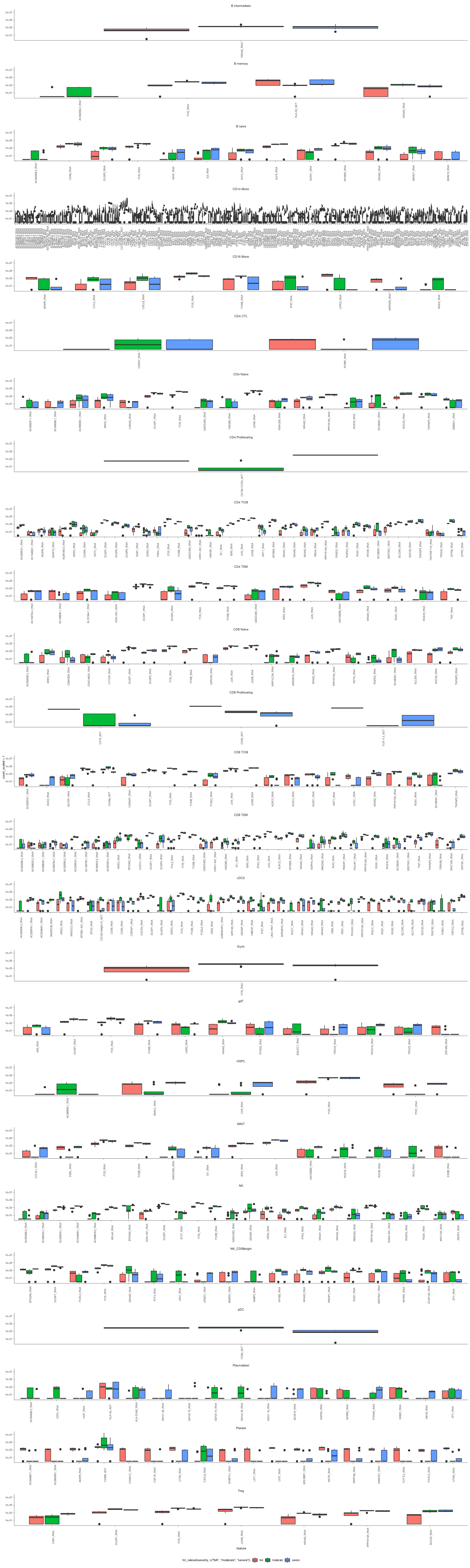
plots_de = readRDS(here("data/3_prime_batch_1/fast_pipeline_results/differential_transcript_abundance/plot_significant.rds"))
Box plots of the differentially abundant gene-transcripts healthy vs moderate or moderate vs severe
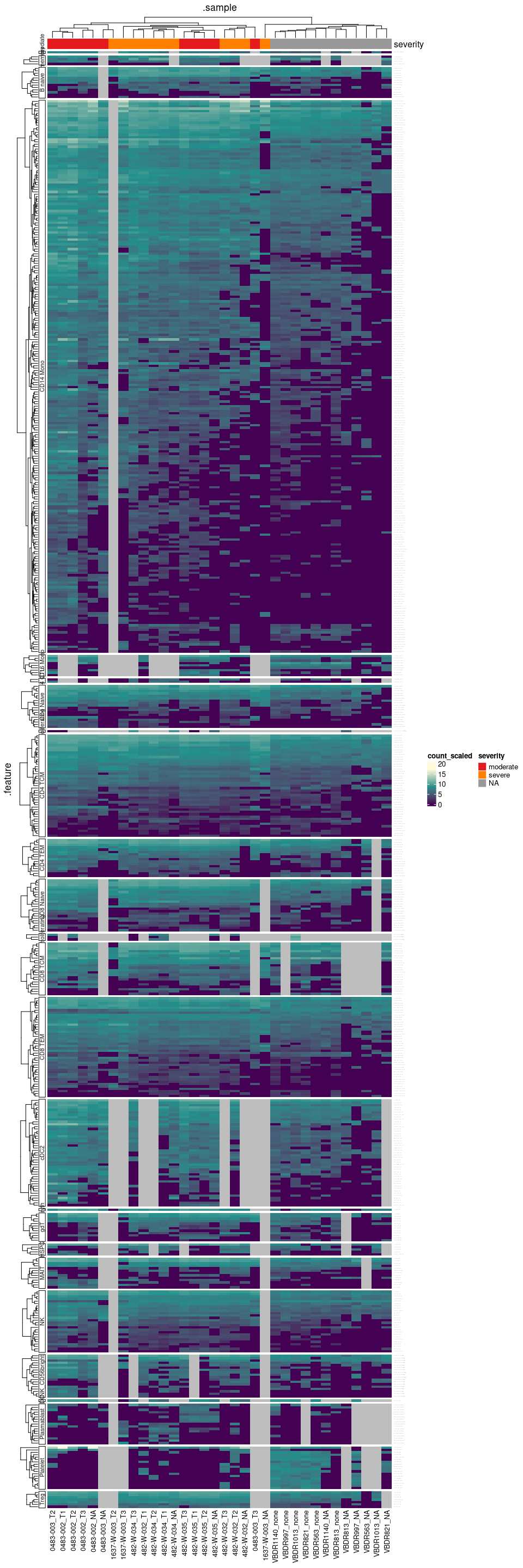
Heatmap of the differentially abundant gene-transcripts healthy vs moderate or moderate vs severe
## Loading required package: tidyHeatmap
## ========================================
## tidyHeatmap version 1.8.1
## If you use tidyHeatmap in published research, please cite:
## 1) Mangiola et al. tidyHeatmap: an R package for modular heatmap production
## based on tidy principles. JOSS 2020.
## 2) Gu, Z. Complex heatmaps reveal patterns and correlations in multidimensional
## genomic data. Bioinformatics 2016.
## This message can be suppressed by:
## suppressPackageStartupMessages(library(tidyHeatmap))
## ========================================
##
## Attaching package: 'tidyHeatmap'
## The following object is masked from 'package:stats':
##
## heatmap
The overlap in significant genes
results_de = readRDS(here("data/3_prime_batch_1/fast_pipeline_results/differential_transcript_abundance/differential_transcript_abundance_output.rds"))
results_de= results_de |> mutate(
significant_genes_1 = map(
se,
~ .x |>
filter(`FDR___severitymoderate-severityNA` < 0.05) |>
distinct(.feature) |>
pull(.feature)
),
significant_genes_2 = map(
se,
~ .x |>
filter(`FDR___severitysevere-severitymoderate` < 0.05) |>
distinct(.feature) |>
pull(.feature)
)
)
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
## tidySummarizedExperiment says: The resulting data frame is not rectangular (all genes for all samples), a tibble is returned for independent data analysis.
## tidySummarizedExperiment says: A data frame is returned for independent data analysis.
Common significant genes for the transition healthy moderate
results_de |>
select(-se) |>
unnest(significant_genes_1) |>
tidybulk:::drop_class("tidySummarizedExperiment_nested") |>
select(-significant_genes_2) |>
nest(data = -significant_genes_1) |>
mutate(n = map_int(data, ~ nrow(.x))) |>
arrange(desc(n)) |>
mutate(cell_types = map_chr(data, ~ .x |> pull(1) |> paste(collapse=", "))) |>
select(-data)
## # A tibble: 332 × 3
## significant_genes_1 n cell_types
## <chr> <int> <chr>
## 1 FOS_RNA 18 B memory, B naive, CD14 Mono, CD16 Mono, CD4 Naive…
## 2 NR4A2_RNA 15 B intermediate, B memory, B naive, CD14 Mono, CD4 …
## 3 DUSP1_RNA 12 CD14 Mono, CD4 Naive, CD4 TCM, CD4 TEM, CD8 Naive,…
## 4 FOSB_RNA 11 CD14 Mono, CD16 Mono, CD4 TCM, CD4 TEM, CD8 Naive,…
## 5 PPP1R15A_RNA 9 CD14 Mono, CD4 Naive, CD4 TCM, CD8 Naive, CD8 TCM,…
## 6 AC004556.3_RNA 7 B memory, B naive, CD14 Mono, CD8 Naive, CD8 TEM, …
## 7 RGS1_RNA 7 CD4 TCM, CD4 TEM, CD8 TCM, CD8 TEM, cDC2, NK, NK_C…
## 8 AC020916.1_RNA 6 CD14 Mono, CD4 TCM, CD8 TCM, CD8 TEM, cDC2, NK
## 9 AREG_RNA 6 CD14 Mono, CD4 Naive, CD4 TCM, CD8 Naive, CD8 TEM,…
## 10 SOCS3_RNA 6 CD14 Mono, CD4 Naive, CD4 TCM, CD8 Naive, cDC2, Tr…
## # … with 322 more rows
Common significant genes for the transition moderate severe
results_de |>
select(-se) |>
unnest(significant_genes_2) |>
tidybulk:::drop_class("tidySummarizedExperiment_nested") |>
select(-significant_genes_1) |>
nest(data = -significant_genes_2) |>
mutate(n = map_int(data, ~ nrow(.x))) |>
arrange(desc(n)) |>
mutate(cell_types = map_chr(data, ~ .x |> pull(1) |> paste(collapse=", "))) |>
select(-data)
## # A tibble: 71 × 3
## significant_genes_2 n cell_types
## <chr> <int> <chr>
## 1 SCGB3A1_RNA 4 CD4 Naive, CD4 TCM, CD8 Naive, CD8 TCM
## 2 KRT1_RNA 2 CD4 TCM, CD8 TCM
## 3 RGPD2_RNA 2 CD4 TCM, CD8 Naive
## 4 AC004556.3_RNA 2 CD8 Naive, Plasmablast
## 5 ZFY_RNA 2 NK_CD56bright, Plasmablast
## 6 IGHE_RNA 1 B naive
## 7 KLRK1_RNA 1 B naive
## 8 CCL4L2_RNA 1 CD14 Mono
## 9 ZFAT_RNA 1 CD14 Mono
## 10 KIAA1324_RNA 1 CD4 Naive
## # … with 61 more rows