Load data
# Load data
pseudobulk =
here("data/3_prime_batch_1/preprocessing_results/pseudobulk_preprocessing/pseudobulk_preprocessing_sample_output.rds") |>
readRDS()
Analysis for RNA
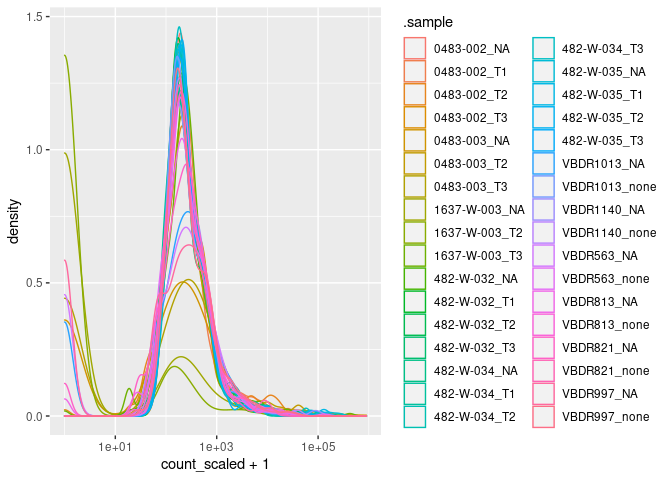
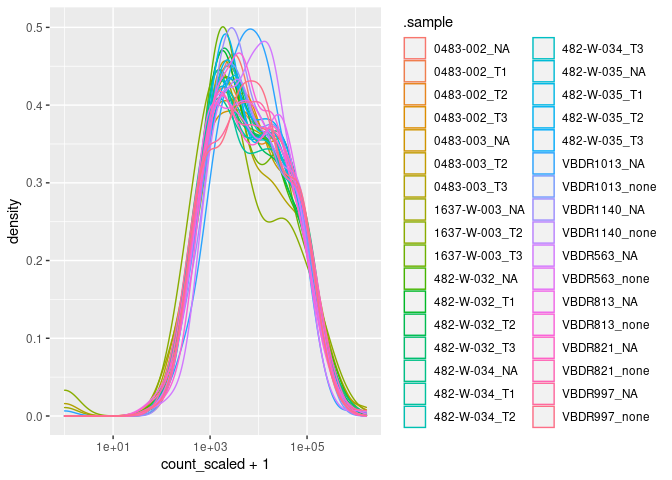
Here to check that the input counts have not global sequencing-depth effect
data_for_pca |>
ggplot(aes(count_scaled + 1, color=.sample)) + geom_density() + scale_x_log10()
Calculate PCA of pseudobulk
metadata =
data_for_pca |>
pivot_sample() |>
select(.sample, donor, simple_timepoint, severity, days_since_symptom_onset, COVID, TMM)
metadata = as.data.frame(metadata)
rownames(metadata) = metadata$`.sample`
metadata = metadata[,-1]
my_pca =
data_for_pca@assays@data$count_scaled |>
log1p() |>
scale() |>
pca(metadata = metadata)
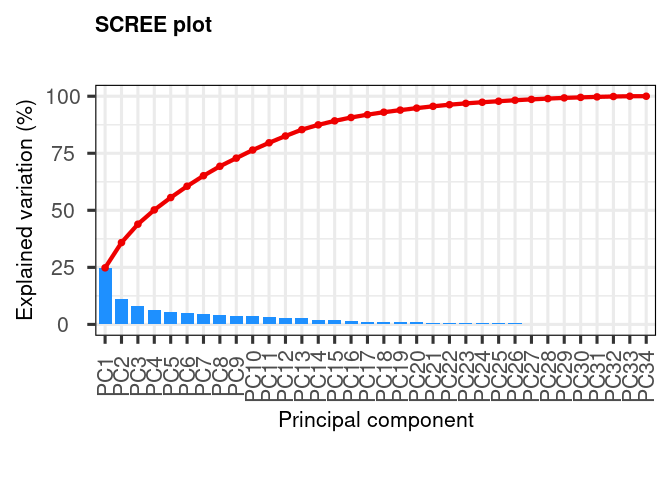
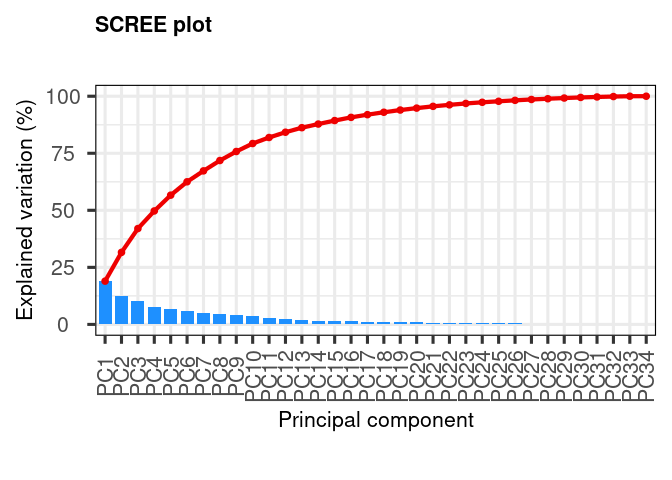
Calculate explained variance of principal components. This gives an idea of how many principal components we need to represent the data faithfully. In this case we see a gradual decrease of variance explained, indicating that we might need up to principal component 20 for explaining 90% of the variance.
my_pca |> screeplot()
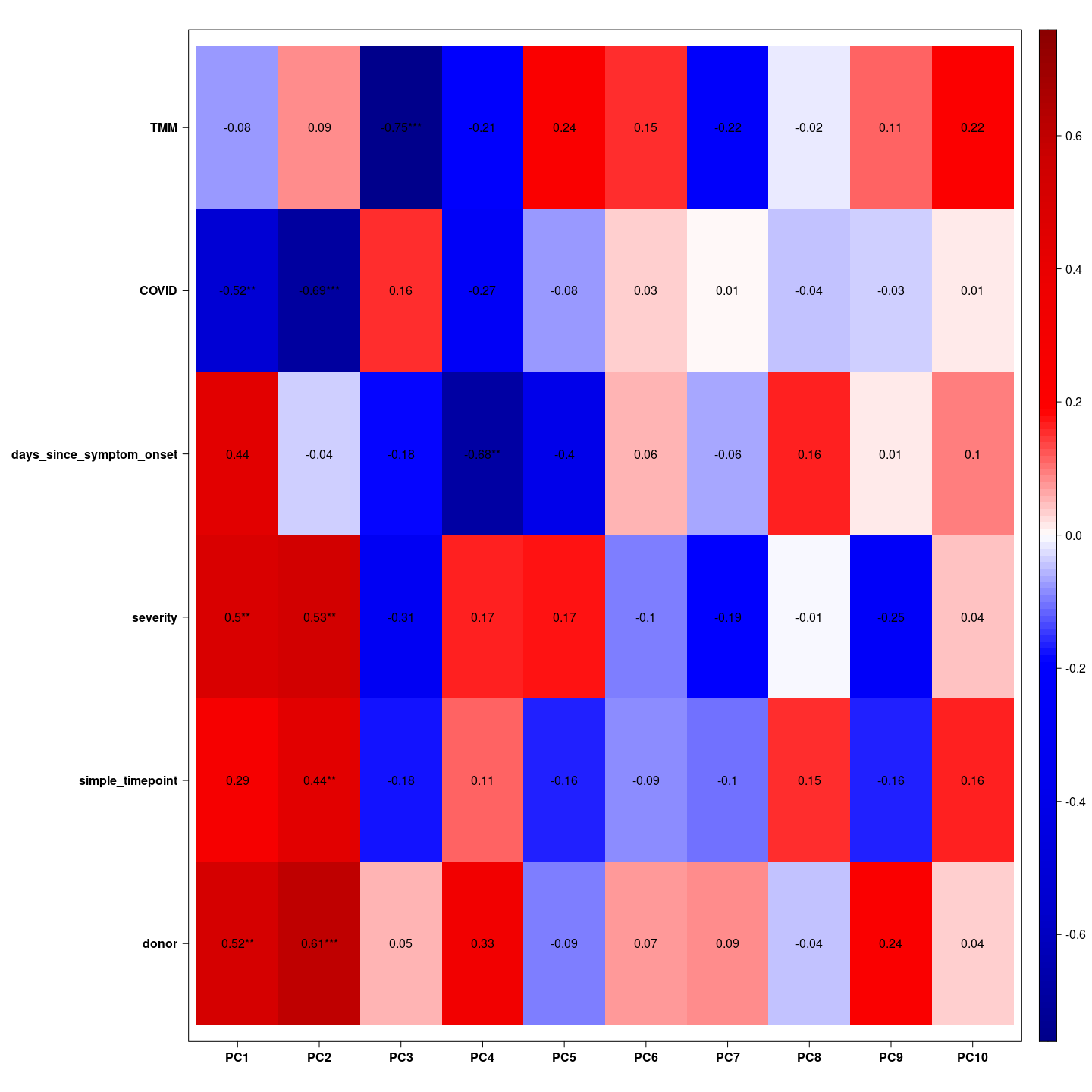
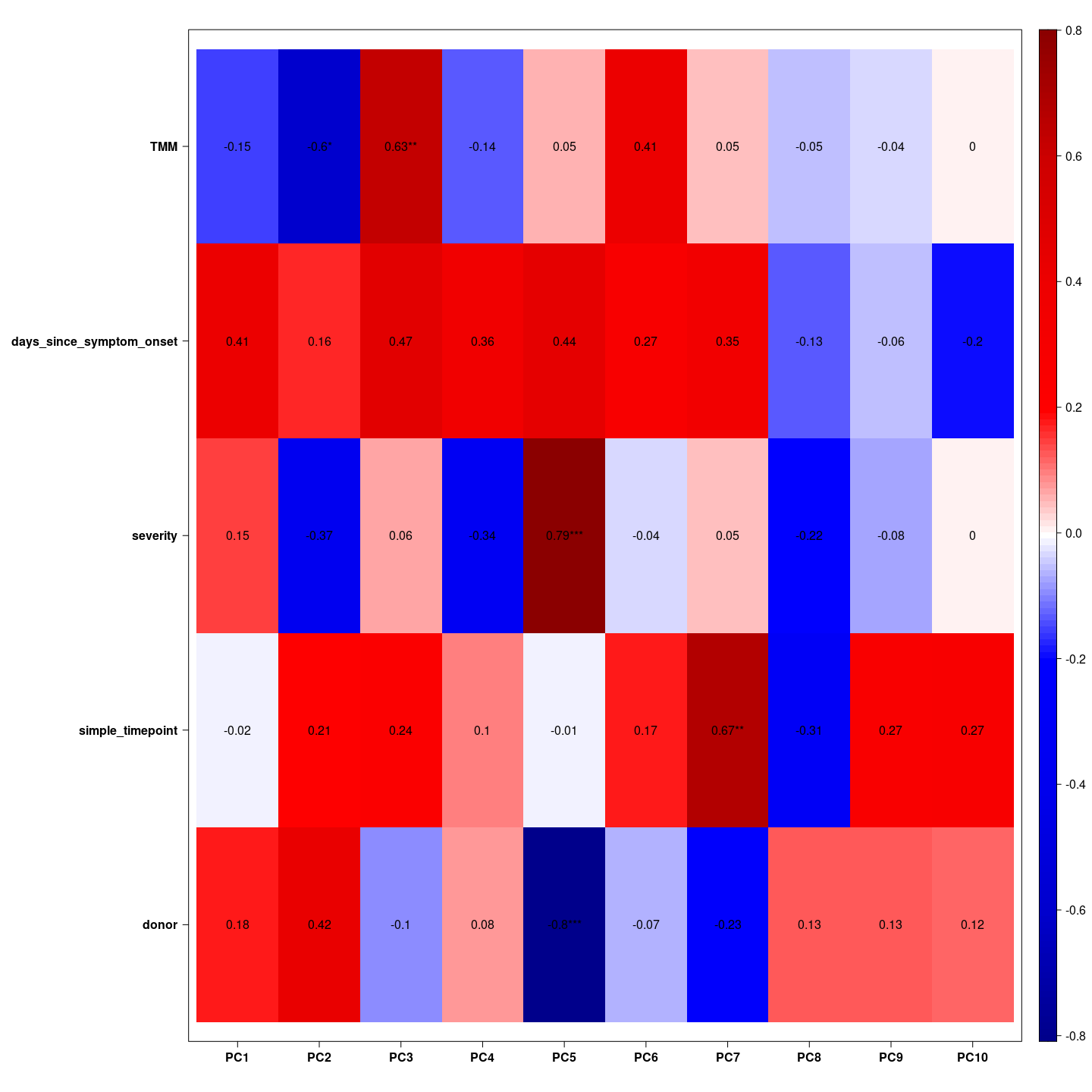
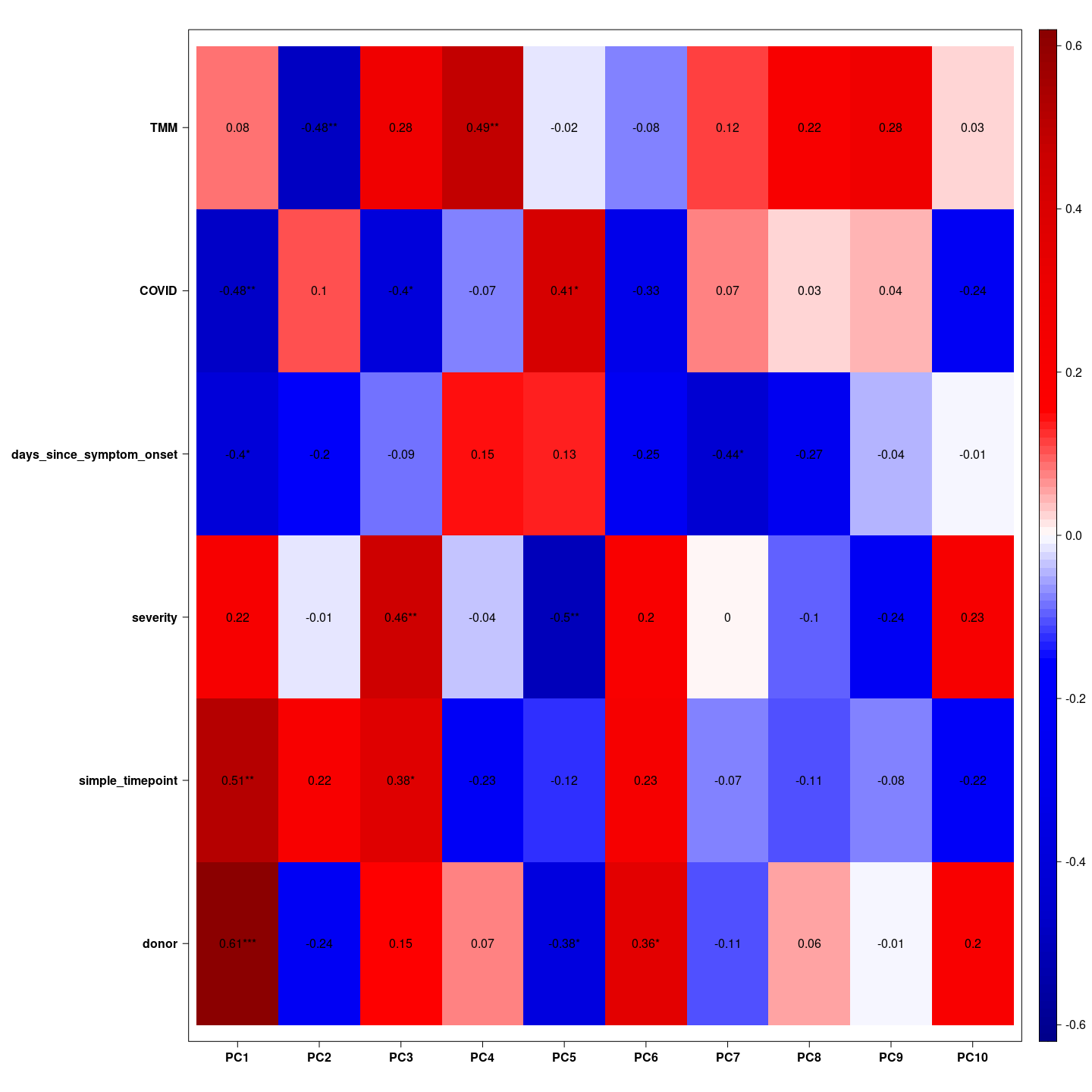
Here we see which variable is associated with which principal component. We hope the biological variable are associated with the top principal components.
= indeed we see COVID and severity associated with the first principal component, COVID and days since symptom onset associated with the second principal component, and TMM associated with the third principal component together with simple time points. These gifts as hope that the biological effects have large magnitude.
my_pca |>
eigencorplot(metavars = colnames(metadata))
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): donor is not
## numeric - please check the source data as non-numeric variables will be coerced
## to numeric
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): simple_timepoint
## is not numeric - please check the source data as non-numeric variables will be
## coerced to numeric
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): severity is not
## numeric - please check the source data as non-numeric variables will be coerced
## to numeric
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): COVID is not
## numeric - please check the source data as non-numeric variables will be coerced
## to numeric
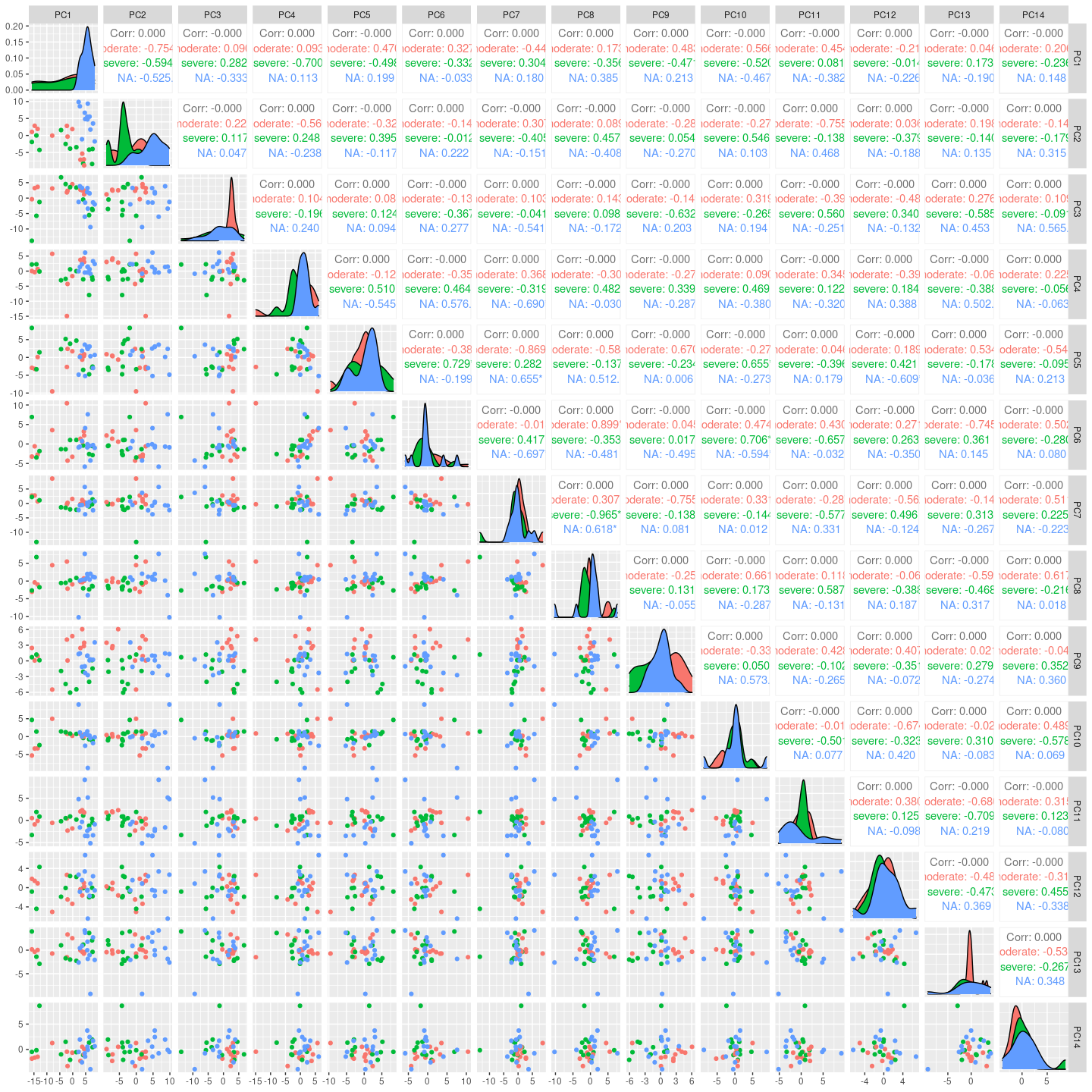
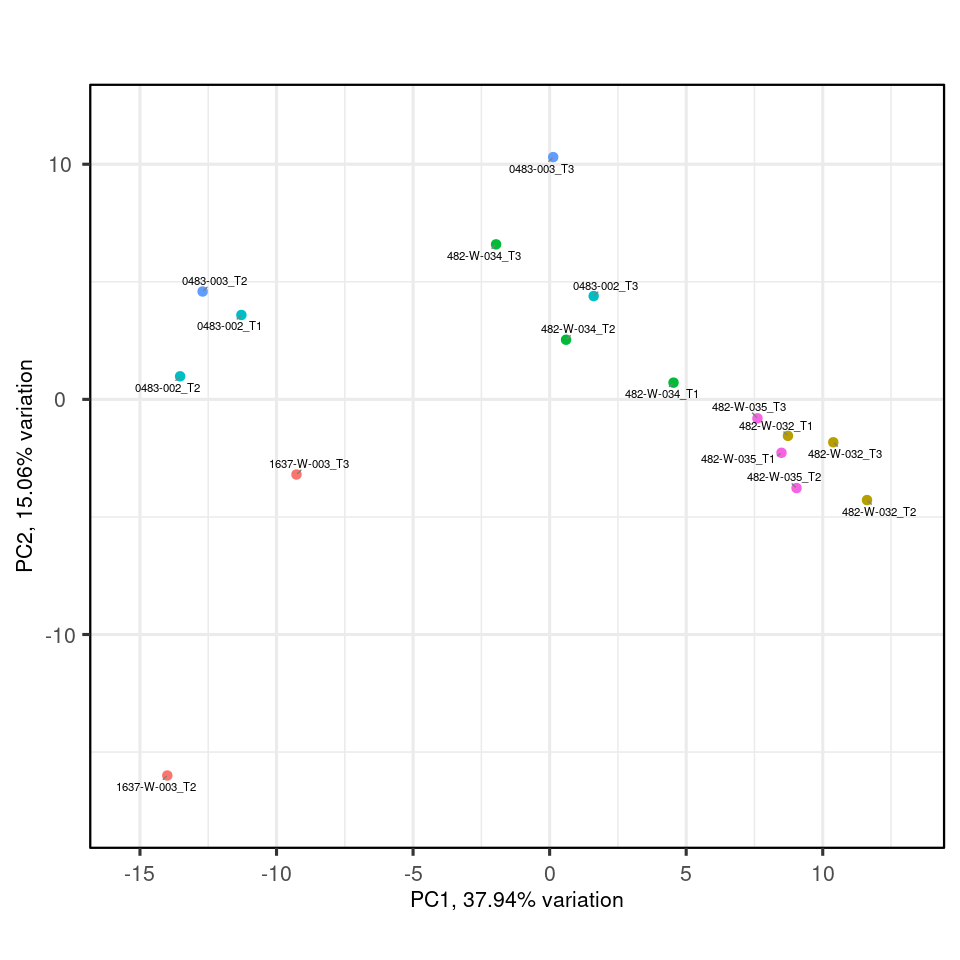
Here we see for the distribution of samples coloured by severity along the top principal components.
my_pca$rotated |>
cbind(my_pca$metadata) |>
as_tibble(rownames = ".sample") |>
GGally::ggpairs(columns = 2:15, ggplot2::aes(colour=`severity`))
## Registered S3 method overwritten by 'GGally':
## method from
## +.gg ggplot2
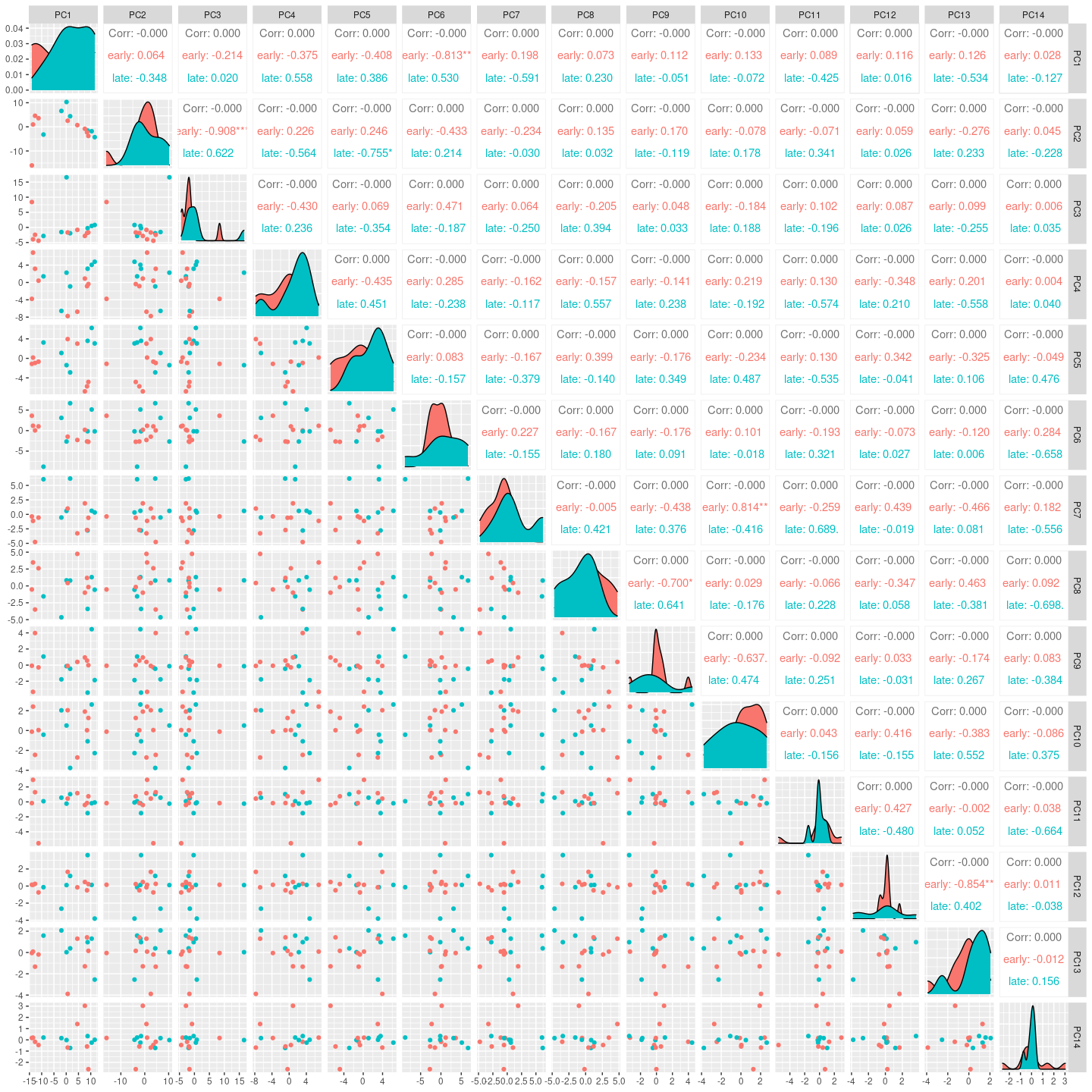
Here we see for the distribution of samples coloured by days_since_symptom_onset along the top principal components.
metadata_with_days =
metadata |>
filter(!is.na(days_since_symptom_onset))
data_for_pca_days =
data_for_pca |>
filter(!is.na(days_since_symptom_onset))
my_pca_days =
filter(data_for_pca_days, !is.na(days_since_symptom_onset))@assays@data$count_scaled |>
log1p() |>
scale() |>
pca(metadata = metadata_with_days)
my_pca_days$rotated |>
cbind(my_pca_days$metadata) |>
as_tibble(rownames = ".sample") |>
mutate(days_since_symptom_onset = if_else(days_since_symptom_onset>median(days_since_symptom_onset), "late", "early")) |>
GGally::ggpairs(columns = 2:15, ggplot2::aes(colour=`days_since_symptom_onset`))
my_pca_days |>
eigencorplot(metavars = colnames(metadata_with_days |> select(-COVID)))
## Warning in eigencorplot(my_pca_days, metavars =
## colnames(select(metadata_with_days, : donor is not numeric - please check the
## source data as non-numeric variables will be coerced to numeric
## Warning in eigencorplot(my_pca_days, metavars =
## colnames(select(metadata_with_days, : simple_timepoint is not numeric - please
## check the source data as non-numeric variables will be coerced to numeric
## Warning in eigencorplot(my_pca_days, metavars =
## colnames(select(metadata_with_days, : severity is not numeric - please check the
## source data as non-numeric variables will be coerced to numeric
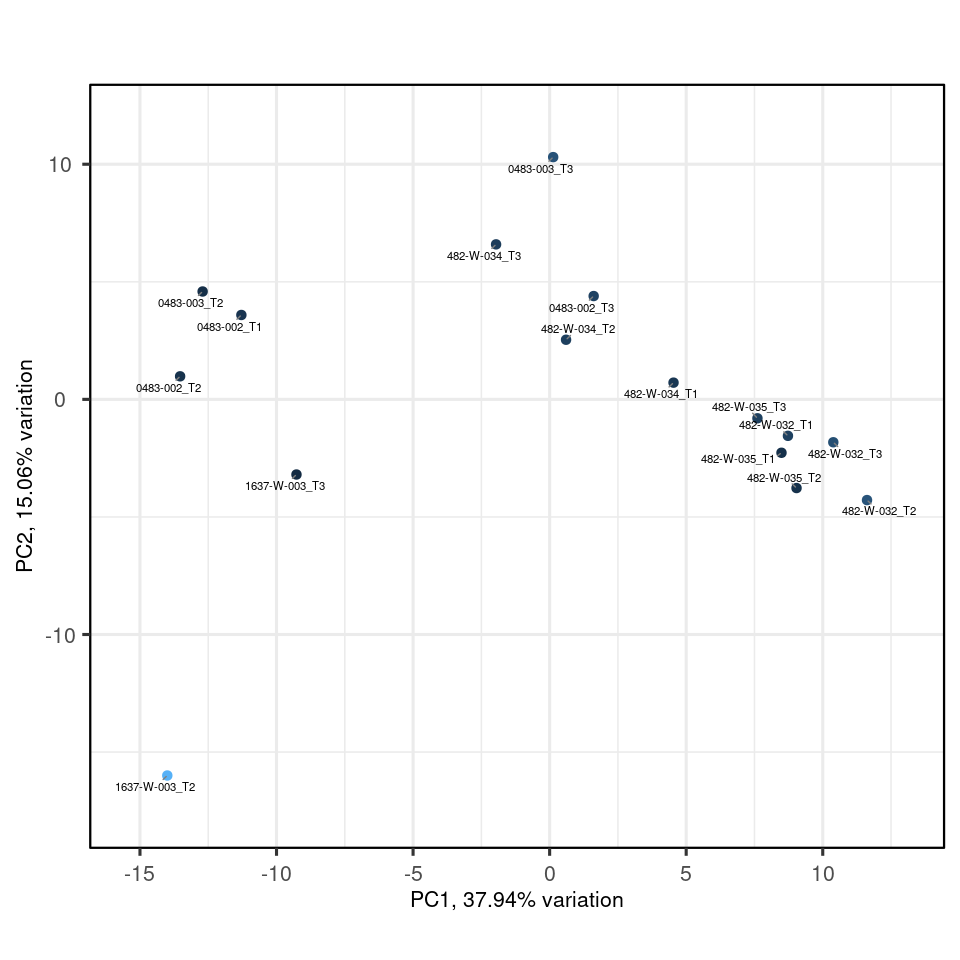
my_pca_days |>
biplot(colby="TMM")
my_pca_days |>
biplot(colby="donor")
Analysis of antibodies
Here to check that the input counts have not global sequencing-depth effect
data_for_pca |>
ggplot(aes(count_scaled + 1, color=.sample)) + geom_density() + scale_x_log10()
Calculate PCA of pseudobulk
metadata =
data_for_pca |>
pivot_sample() |>
select(.sample, donor, simple_timepoint, severity, days_since_symptom_onset, COVID, TMM) |>
replace_na(replace = list(days_since_symptom_onset = 0))
metadata = as.data.frame(metadata)
rownames(metadata) = metadata$`.sample`
metadata = metadata[,-1]
my_pca =
data_for_pca@assays@data$count_scaled |>
log1p() |>
scale() |>
pca(metadata = metadata)
Calculate explained variance of principal components. This gives an idea of how many principal components we need to represent the data faithfully. In this case we see a gradual decrease of variance explained, indicating that we might need up to printipal component 20 for explaining 90% of the variance.
my_pca |> screeplot()
 Here we see which variable is associated with which principal component.
We hope the biological variable are associated with the top principal
components.
Here we see which variable is associated with which principal component.
We hope the biological variable are associated with the top principal
components.
= indeed we see COVID and severity associated with the first principal component, COVID and days since symptom onset associated with the second principal component, and TMM associated with the third principal component together with simple time points. These gifts as hope that the biological effects have large magnitude.
my_pca |>
eigencorplot(metavars = colnames(metadata))
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): donor is not
## numeric - please check the source data as non-numeric variables will be coerced
## to numeric
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): simple_timepoint
## is not numeric - please check the source data as non-numeric variables will be
## coerced to numeric
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): severity is not
## numeric - please check the source data as non-numeric variables will be coerced
## to numeric
## Warning in eigencorplot(my_pca, metavars = colnames(metadata)): COVID is not
## numeric - please check the source data as non-numeric variables will be coerced
## to numeric
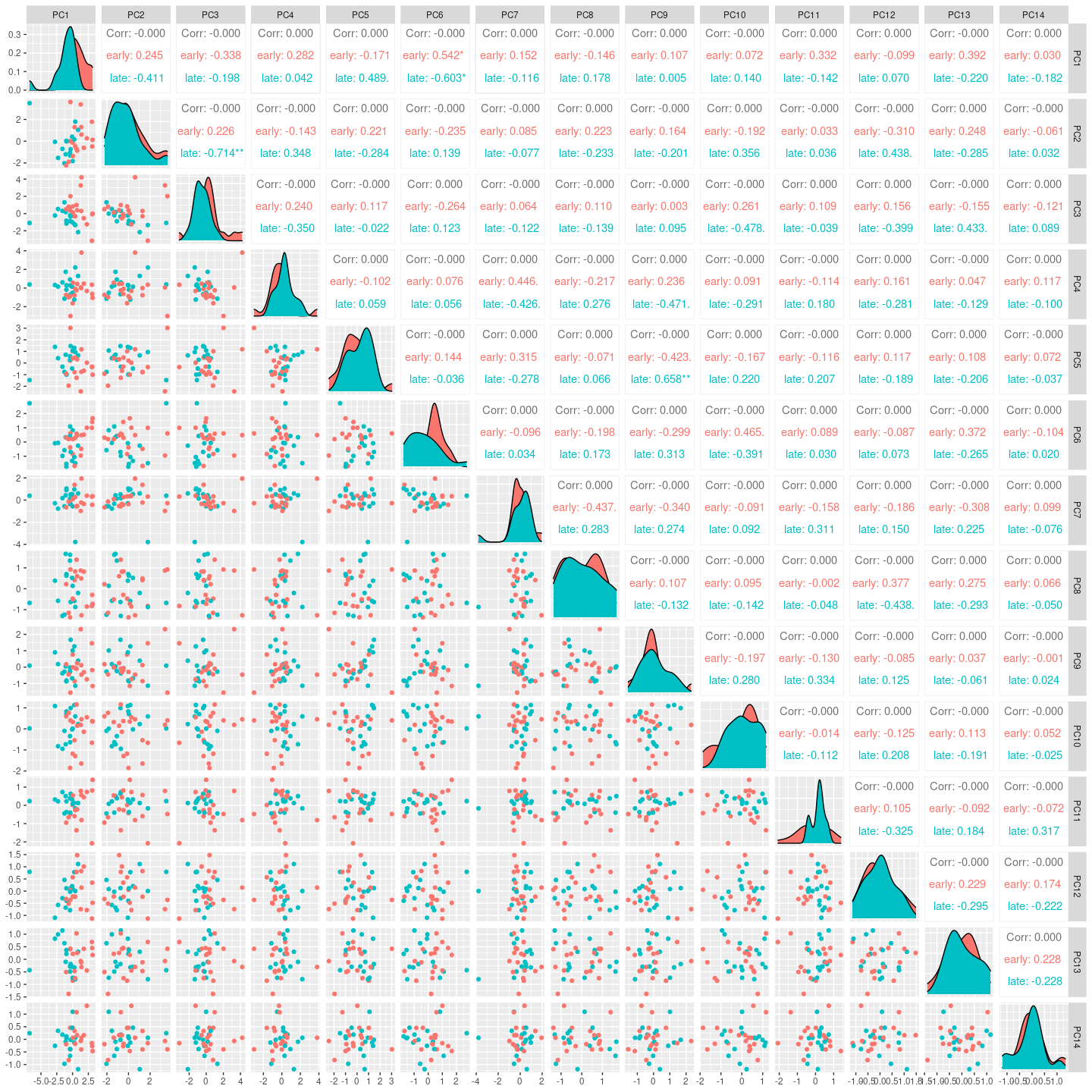
 Here we see for the distribution of samples coloused by severity along
the top principal components.
Here we see for the distribution of samples coloused by severity along
the top principal components.
my_pca$rotated |>
cbind(my_pca$metadata) |>
as_tibble(rownames = ".sample") |>
GGally::ggpairs(columns = 2:15, ggplot2::aes(colour=`severity`))
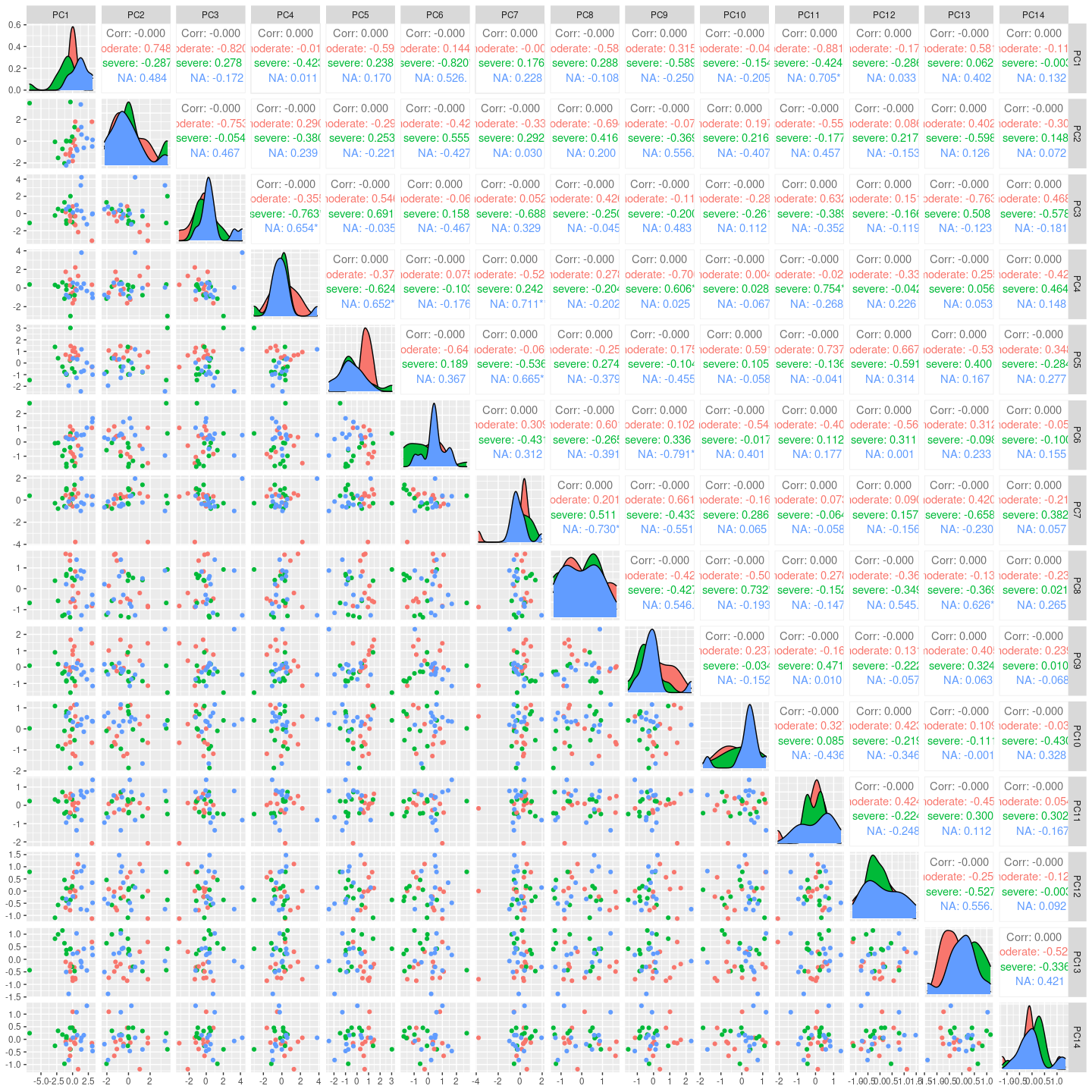 Here we see for the distribution of samples coloused by
days_since_symptom_onset along the top principal components.
Here we see for the distribution of samples coloused by
days_since_symptom_onset along the top principal components.
my_pca$rotated |>
cbind(my_pca$metadata) |>
as_tibble(rownames = ".sample") |>
mutate(days_since_symptom_onset = if_else(days_since_symptom_onset>median(days_since_symptom_onset), "late", "early")) |>
GGally::ggpairs(columns = 2:15, ggplot2::aes(colour=`days_since_symptom_onset`))
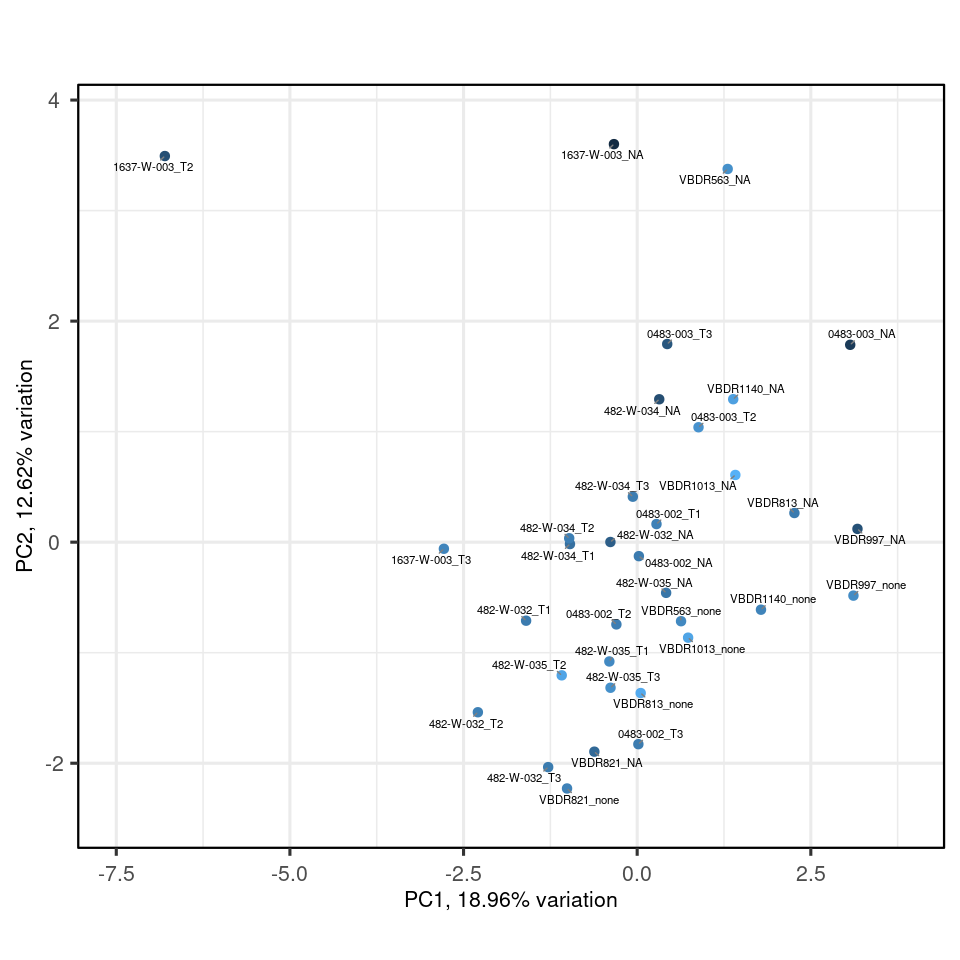
my_pca |>
biplot(colby="TMM")
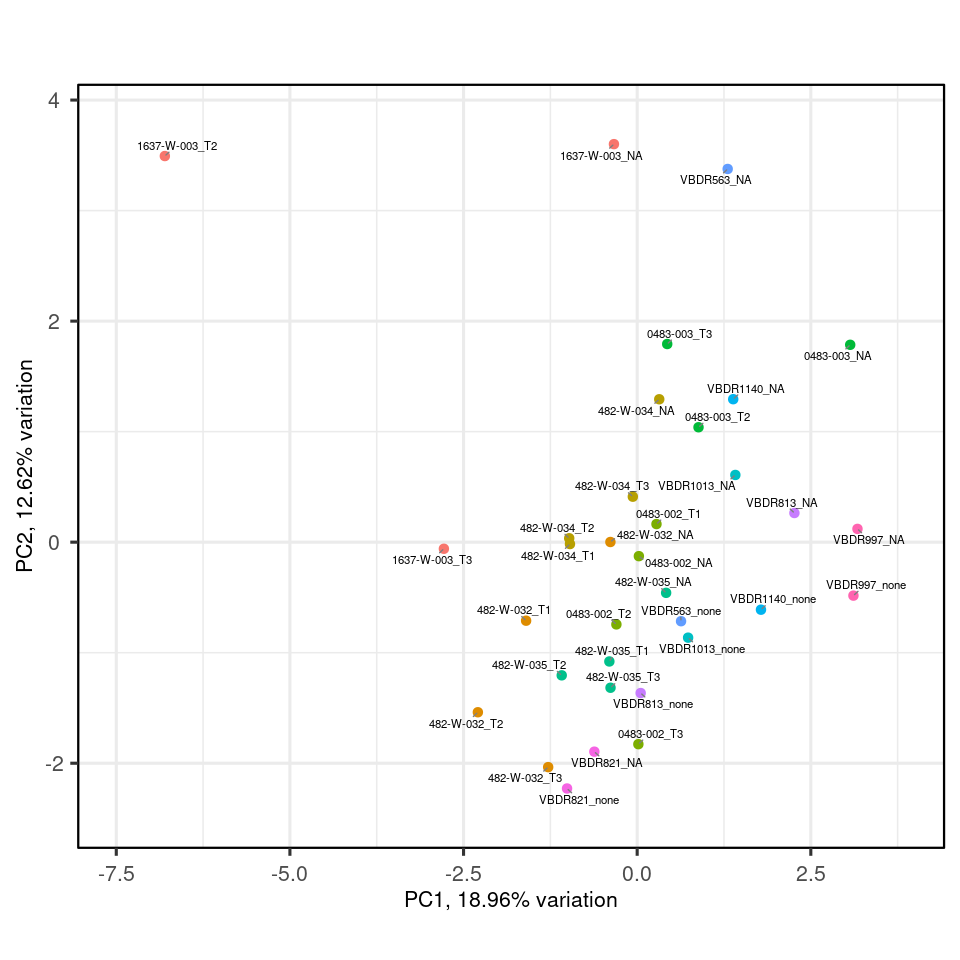
my_pca |>
biplot(colby="donor")
Analysis of covariate redundancy
metadata
## donor simple_timepoint severity days_since_symptom_onset
## 0483-002_NA 0483-002 NA moderate 0
## 0483-002_T1 0483-002 T1 moderate 5
## 0483-002_T2 0483-002 T2 moderate 8
## 0483-002_T3 0483-002 T3 moderate 34
## 0483-003_NA 0483-003 NA moderate 0
## 0483-003_T2 0483-003 T2 moderate 7
## 0483-003_T3 0483-003 T3 moderate 39
## 1637-W-003_NA 1637-W-003 NA severe 0
## 1637-W-003_T2 1637-W-003 T2 severe 11
## 1637-W-003_T3 1637-W-003 T3 severe 17
## 482-W-032_NA 482-W-032 NA severe 0
## 482-W-032_T1 482-W-032 T1 severe 33
## 482-W-032_T2 482-W-032 T2 severe 37
## 482-W-032_T3 482-W-032 T3 severe 39
## 482-W-034_NA 482-W-034 NA severe 0
## 482-W-034_T1 482-W-034 T1 severe 9
## 482-W-034_T2 482-W-034 T2 severe 11
## 482-W-034_T3 482-W-034 T3 severe 15
## 482-W-035_NA 482-W-035 NA moderate 0
## 482-W-035_T1 482-W-035 T1 moderate 3
## 482-W-035_T2 482-W-035 T2 moderate 5
## 482-W-035_T3 482-W-035 T3 moderate 9
## VBDR1013_NA VBDR1013 NA NA 0
## VBDR1013_none VBDR1013 none NA 0
## VBDR1140_NA VBDR1140 NA NA 0
## VBDR1140_none VBDR1140 none NA 0
## VBDR563_NA VBDR563 NA NA 0
## VBDR563_none VBDR563 none NA 0
## VBDR813_NA VBDR813 NA NA 0
## VBDR813_none VBDR813 none NA 0
## VBDR821_NA VBDR821 NA NA 0
## VBDR821_none VBDR821 none NA 0
## VBDR997_NA VBDR997 NA NA 0
## VBDR997_none VBDR997 none NA 0
## COVID TMM
## 0483-002_NA TRUE 1.0074424
## 0483-002_T1 TRUE 1.0255025
## 0483-002_T2 TRUE 1.0156813
## 0483-002_T3 TRUE 1.0106802
## 0483-003_NA TRUE 0.7986725
## 0483-003_T2 TRUE 1.0774194
## 0483-003_T3 TRUE 0.8916291
## 1637-W-003_NA TRUE 0.7399618
## 1637-W-003_T2 TRUE 0.8664717
## 1637-W-003_T3 TRUE 1.0360248
## 482-W-032_NA TRUE 0.9160799
## 482-W-032_T1 TRUE 1.0076738
## 482-W-032_T2 TRUE 1.0242726
## 482-W-032_T3 TRUE 1.0082499
## 482-W-034_NA TRUE 0.8628639
## 482-W-034_T1 TRUE 0.9733934
## 482-W-034_T2 TRUE 1.0188914
## 482-W-034_T3 TRUE 1.0121775
## 482-W-035_NA TRUE 0.9916354
## 482-W-035_T1 TRUE 1.0499111
## 482-W-035_T2 TRUE 1.1296713
## 482-W-035_T3 TRUE 1.0691957
## VBDR1013_NA FALSE 1.1674736
## VBDR1013_none FALSE 1.1313864
## VBDR1140_NA FALSE 1.1174852
## VBDR1140_none FALSE 1.0329846
## VBDR563_NA FALSE 1.0694319
## VBDR563_none FALSE 1.0488852
## VBDR813_NA FALSE 1.0065996
## VBDR813_none FALSE 1.1475235
## VBDR821_NA FALSE 0.9507530
## VBDR821_none FALSE 1.0278402
## VBDR997_NA FALSE 0.8730673
## VBDR997_none FALSE 1.0570607
require(tidyverse)
require(rcompanion)
## Loading required package: rcompanion
require(corrr)
## Loading required package: corrr
##
## Attaching package: 'corrr'
## The following object is masked from 'package:tidybulk':
##
## as_matrix
# Calculate a pairwise association between all variables in a data-frame. In particular nominal vs nominal with Chi-square, numeric vs numeric with Pearson correlation, and nominal vs numeric with ANOVA.
# Adopted from https://stackoverflow.com/a/52557631/590437
mixed_assoc = function(df, cor_method="spearman", adjust_cramersv_bias=TRUE){
# https://stackoverflow.com/questions/52554336/plot-the-equivalent-of-correlation-matrix-for-factors-categorical-data-and-mi
df_comb = expand.grid(names(df), names(df), stringsAsFactors = F) %>% set_names(c("X1", "X2"))
is_nominal = function(x) class(x) %in% c("factor", "character")
# https://community.rstudio.com/t/why-is-purr-is-numeric-deprecated/3559
# https://github.com/r-lib/rlang/issues/781
is_numeric <- function(x) { is.integer(x) || is_double(x)}
f = function(xName,yName) {
x = pull(df, xName)
y = pull(df, yName)
result = if(is_nominal(x) && is_nominal(y)){
# use bias corrected cramersV as described in https://rdrr.io/cran/rcompanion/man/cramerV.html
cv = cramerV(as.character(x), as.character(y), bias.correct = adjust_cramersv_bias)
data.frame(xName, yName, assoc=cv, type="cramersV")
}else if(is_numeric(x) && is_numeric(y)){
correlation = cor(x, y, method=cor_method, use="complete.obs")
data.frame(xName, yName, assoc=correlation, type="correlation")
}else if(is_numeric(x) && is_nominal(y)){
# from https://stats.stackexchange.com/questions/119835/correlation-between-a-nominal-iv-and-a-continuous-dv-variable/124618#124618
r_squared = summary(lm(x ~ y))$r.squared
data.frame(xName, yName, assoc=sqrt(r_squared), type="anova")
}else if(is_nominal(x) && is_numeric(y)){
r_squared = summary(lm(y ~x))$r.squared
data.frame(xName, yName, assoc=sqrt(r_squared), type="anova")
}else {
warning(paste("unmatched column type combination: ", class(x), class(y)))
return(NULL)
}
# finally add complete obs number and ratio to table
result %>% mutate(complete_obs_pairs=sum(!is.na(x) & !is.na(y)), complete_obs_ratio=complete_obs_pairs/length(x)) %>% rename(x=xName, y=yName)
}
# apply function to each variable combination
map2_df(df_comb$X1, df_comb$X2, f)
}
mixed_assoc(metadata)
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## factor
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## factor
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## factor
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## numeric
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: factor
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: factor
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: factor
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: numeric
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: numeric
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## numeric
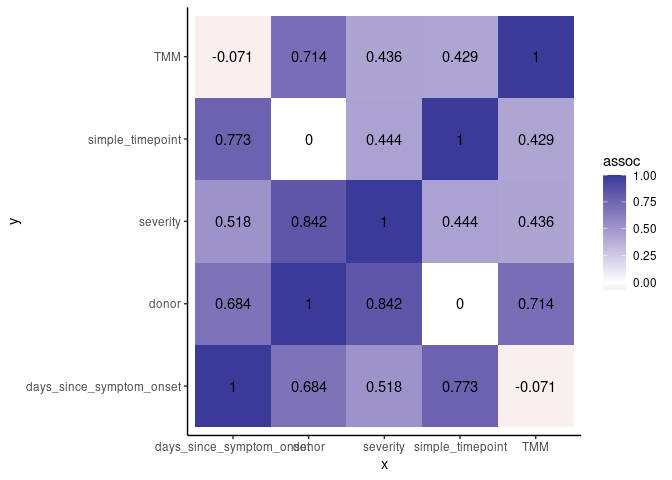
## x y assoc
## Cramer V...1 donor donor 1.00000000
## Cramer V...2 simple_timepoint donor 0.00000000
## Cramer V...3 severity donor 0.84240000
## ...4 days_since_symptom_onset donor 0.68386487
## ...5 TMM donor 0.71400293
## Cramer V...6 donor simple_timepoint 0.00000000
## Cramer V...7 simple_timepoint simple_timepoint 1.00000000
## Cramer V...8 severity simple_timepoint 0.44450000
## ...9 days_since_symptom_onset simple_timepoint 0.77262321
## ...10 TMM simple_timepoint 0.42901040
## Cramer V...11 donor severity 0.84240000
## Cramer V...12 simple_timepoint severity 0.44450000
## Cramer V...13 severity severity 1.00000000
## ...14 days_since_symptom_onset severity 0.51794104
## ...15 TMM severity 0.43571360
## ...16 donor days_since_symptom_onset 0.68386487
## ...17 simple_timepoint days_since_symptom_onset 0.77262321
## ...18 severity days_since_symptom_onset 0.51794104
## ...19 days_since_symptom_onset days_since_symptom_onset 1.00000000
## ...20 TMM days_since_symptom_onset -0.07103901
## ...21 donor TMM 0.71400293
## ...22 simple_timepoint TMM 0.42901040
## ...23 severity TMM 0.43571360
## ...24 days_since_symptom_onset TMM -0.07103901
## ...25 TMM TMM 1.00000000
## type complete_obs_pairs complete_obs_ratio
## Cramer V...1 cramersV 34 1
## Cramer V...2 cramersV 34 1
## Cramer V...3 cramersV 34 1
## ...4 anova 34 1
## ...5 anova 34 1
## Cramer V...6 cramersV 34 1
## Cramer V...7 cramersV 34 1
## Cramer V...8 cramersV 34 1
## ...9 anova 34 1
## ...10 anova 34 1
## Cramer V...11 cramersV 34 1
## Cramer V...12 cramersV 34 1
## Cramer V...13 cramersV 34 1
## ...14 anova 34 1
## ...15 anova 34 1
## ...16 anova 34 1
## ...17 anova 34 1
## ...18 anova 34 1
## ...19 correlation 34 1
## ...20 correlation 34 1
## ...21 anova 34 1
## ...22 anova 34 1
## ...23 anova 34 1
## ...24 correlation 34 1
## ...25 correlation 34 1
Heatmap of correlation
metadata %>%
mixed_assoc() %>%
mutate(assoc = round(assoc, 3)) |>
select(x, y, assoc) %>%
ggplot(aes(x,y,fill=assoc))+
geom_tile()+
geom_text(aes(x,y,label=assoc))+
scale_fill_gradient2() +
theme_classic()
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## factor
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## factor
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## factor
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## numeric
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: factor
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: factor
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: factor
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: numeric
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: numeric
## logical
## Warning in .f(.x[[i]], .y[[i]], ...): unmatched column type combination: logical
## numeric
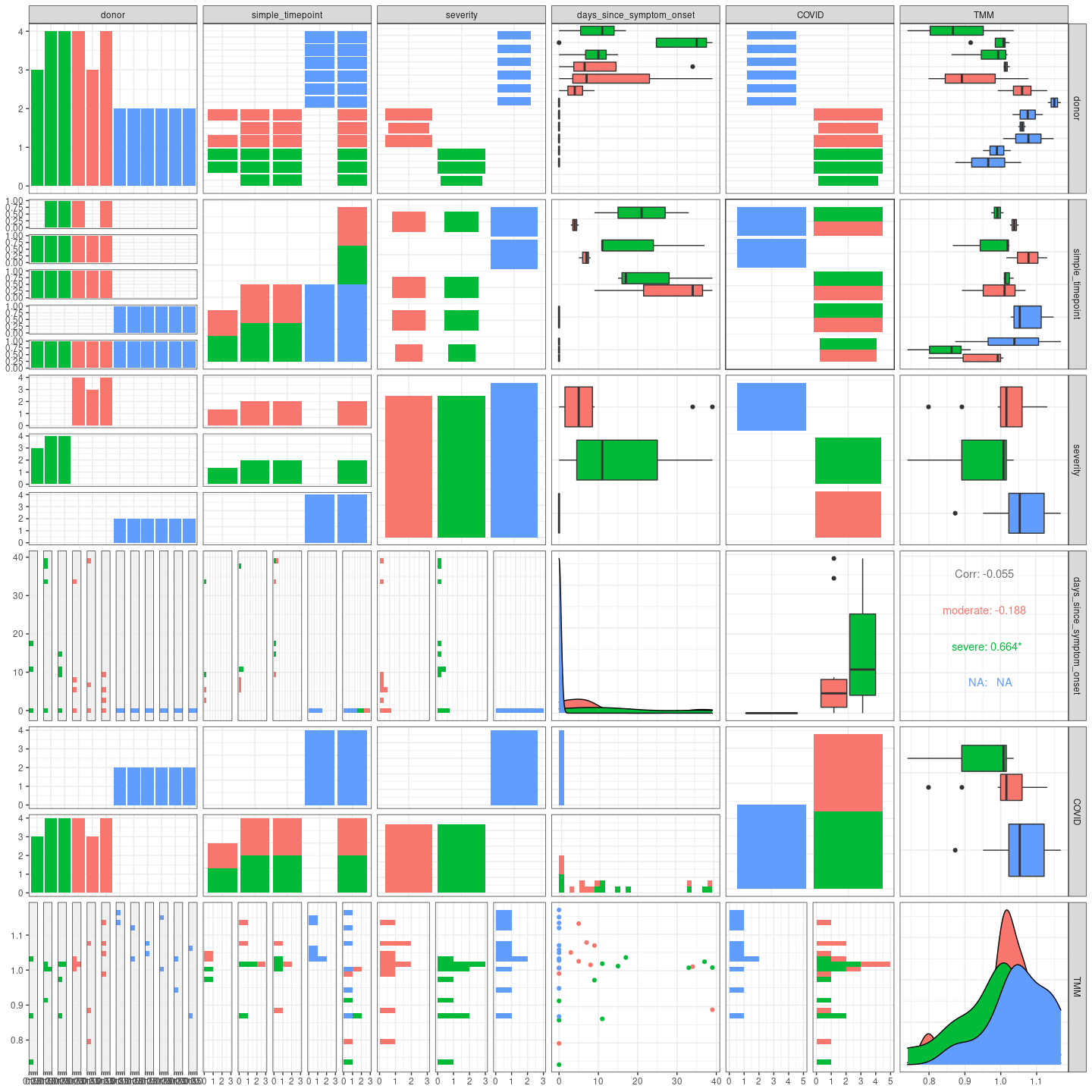
Pair plot, all vs all.
GGally::ggpairs(metadata, aes(color = severity)) + theme_bw()
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## Warning in cor(x, y): the standard deviation is zero
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.